Estimating RNA modifications using Nanopore data
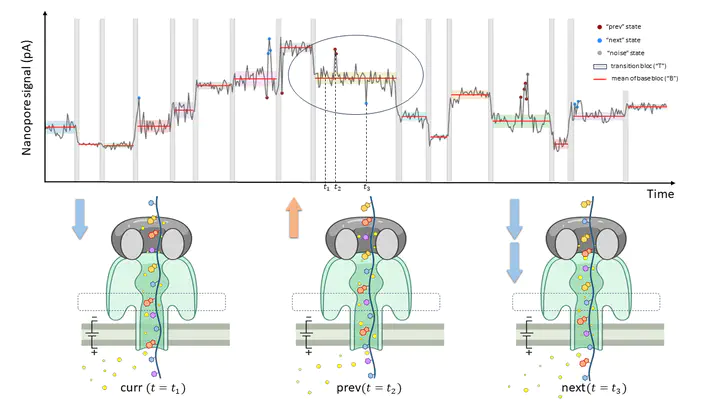
SegPore is a software to segment the raw Nanopore direct RNA sequence and to estimate various RNA modifications and secondary folding structures.
Oxford Nanopore (ONT) is the third generation sequencing technology. It measures the electric current when a RNA molecule passes through the pore. In principle, different signals are generated for different nucleotides. Current computational methods could not provide satisfactory results due to the inaccurate segmentation of the raw signal. We develop a new method, SegPore, that utilizes a molecular jiggling translocation hypothesis to segment the raw signal. Based on the signal segmentation, we demonstrate the interpretable results and decent performances on m6A and inosine modification estimation, as well as RNA secondary structure estimation of SegPore. We find that the end points of the reads take place at the start of estimated stem structures in the reverse transcription direction. Our results indicate the SegPore has the capability to concurrently estimate multiple modifications at the individual molecule level using the same Nanopore direct RNA sequencing data. Additionally, our study reveals novel perspectives for RNA structure estimation.
Preprint: https://www.biorxiv.org/content/10.1101/2024.01.11.575207v1